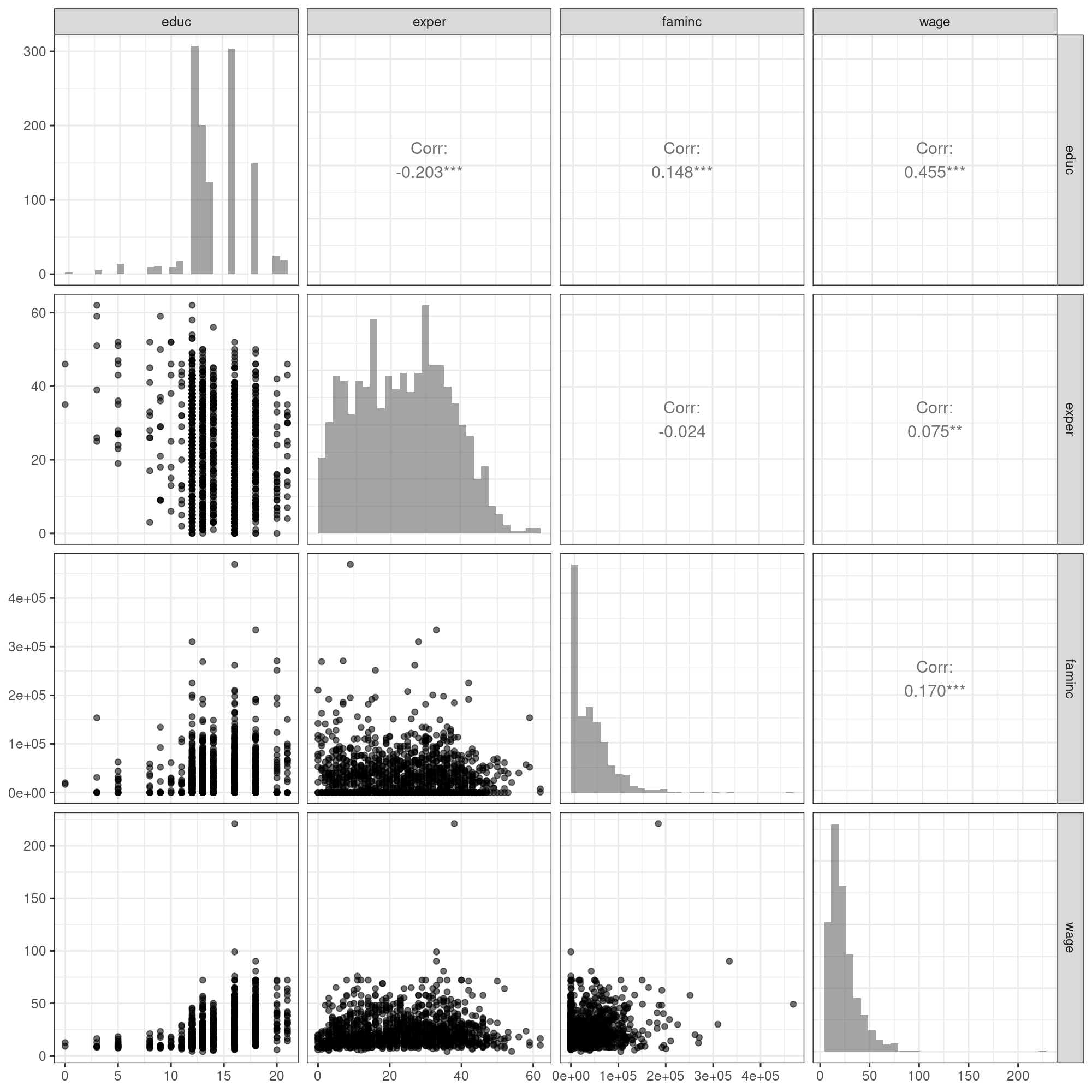
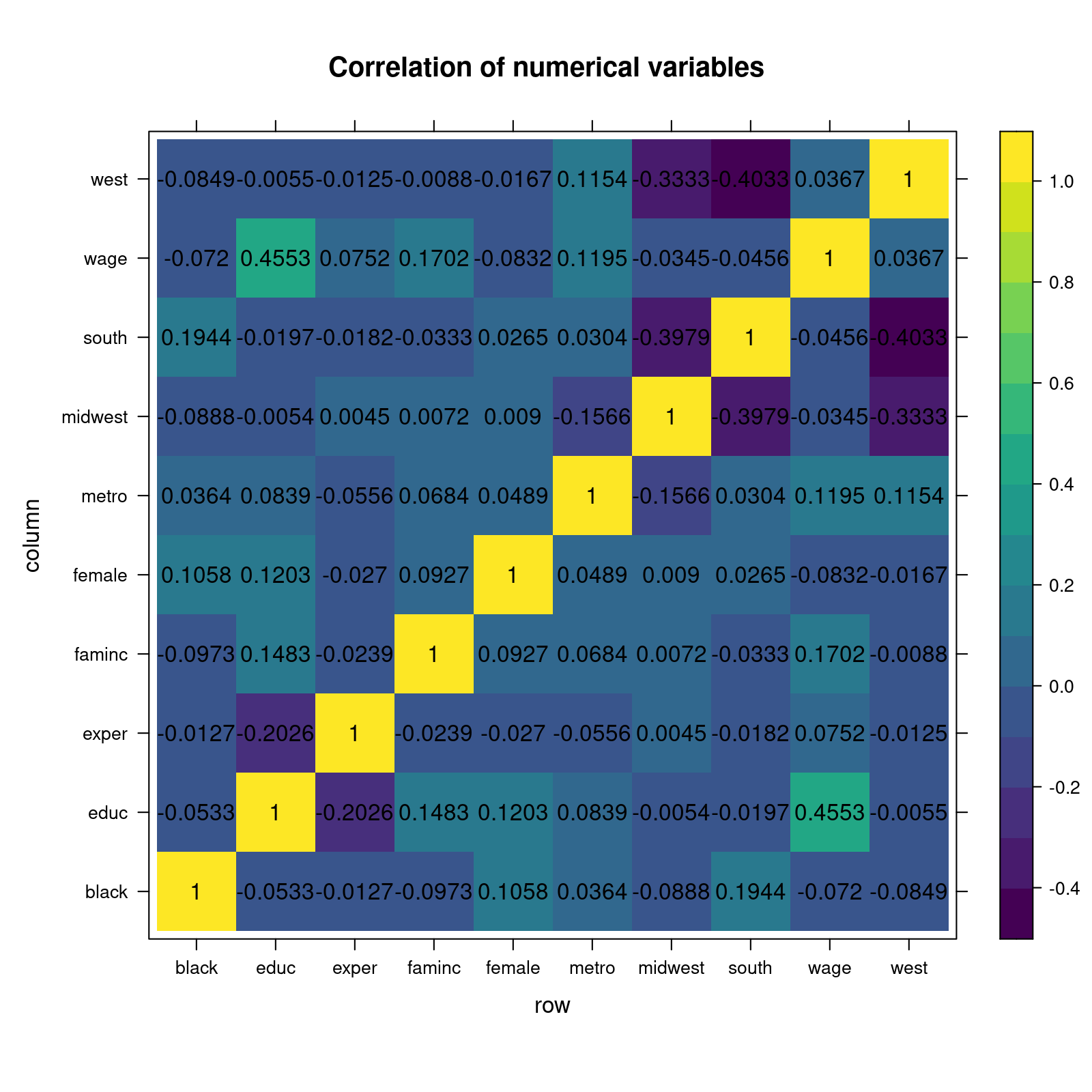
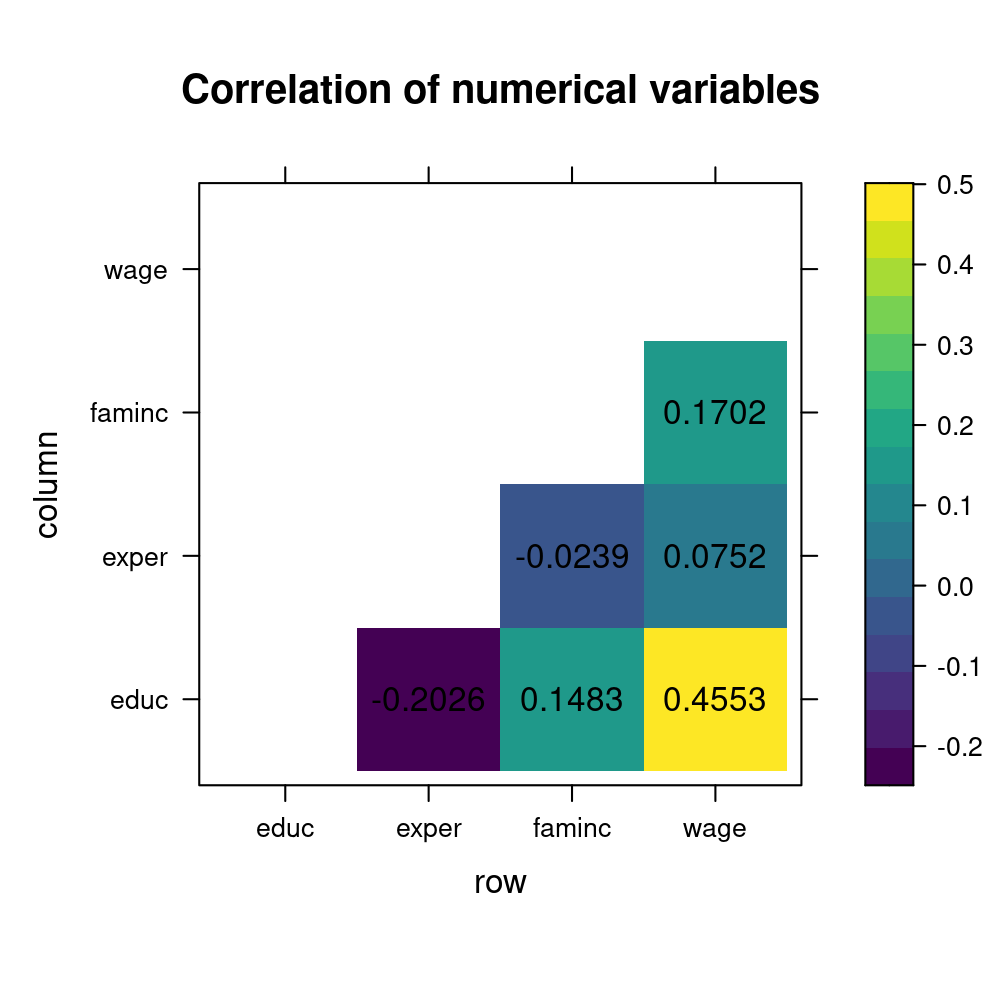
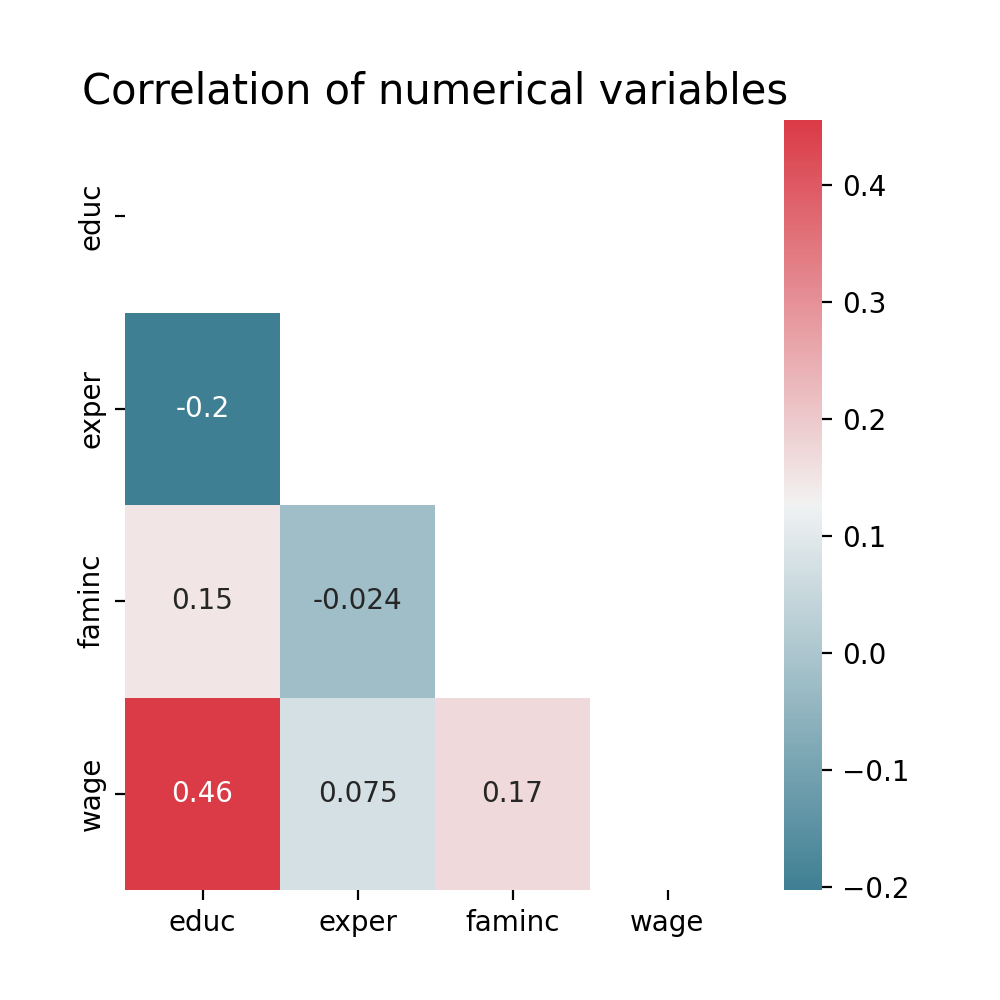
Rows: 1,200
Columns: 10
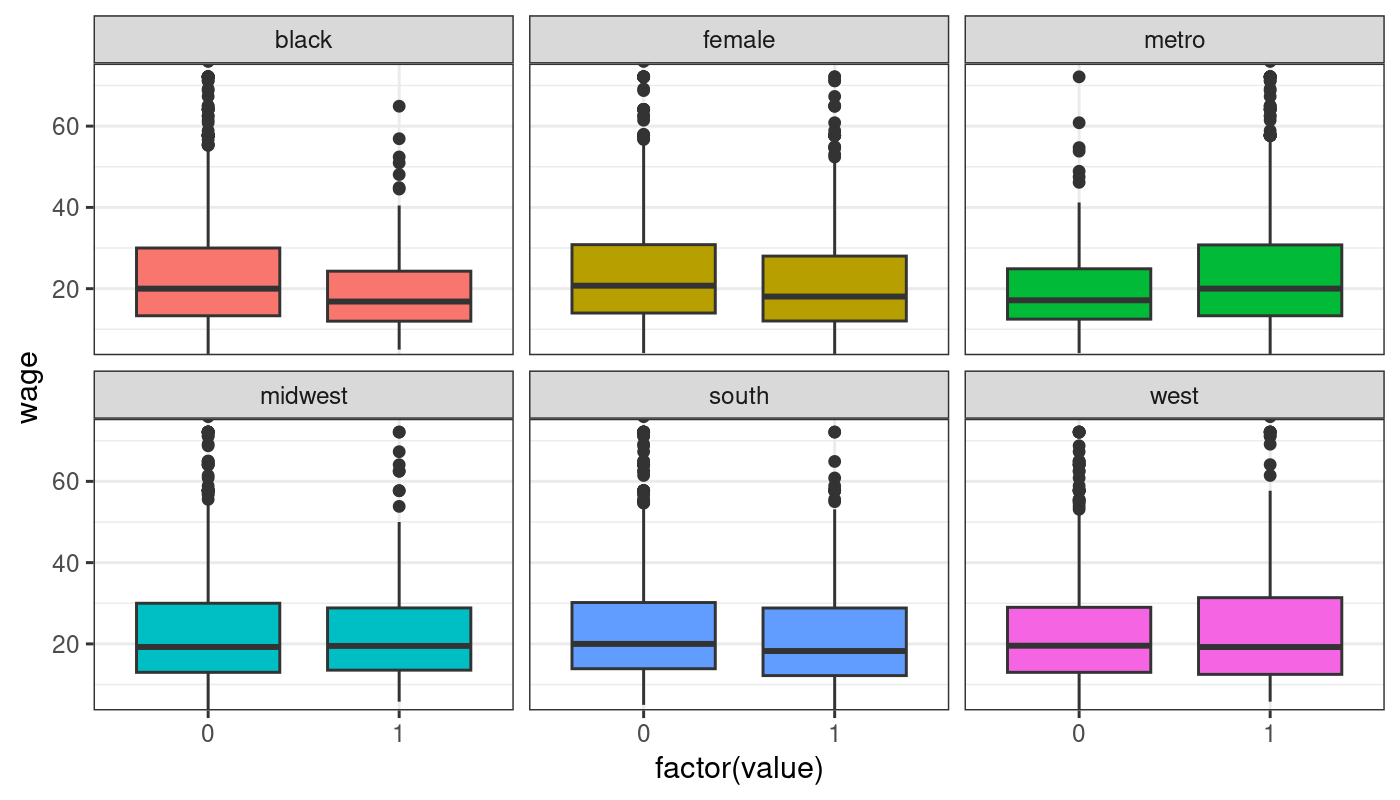
$ black <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, …
$ educ <int> 13, 14, 18, 13, 13, 16, 16, 18, 21, 14, 18, 12, 12, 12, 16, 16, 16, 16, 20, 14, 12, 12, 14, 12, 12, 12, 8, 12, 12, 14, 18, 16, 12, 18, 18, 18, 14, 13, 12, 12, 12, 16, 12, 12, 12, 12, 12, 13, 13, 13, 12, 12, 12, 16, 13, 16, 16, 12, 20, 16, 14, 16, 12, 14, 12, 16, 12, 21, 16, 18, 18, 13, 16, 12, 13, 21, 12, 14, 18, 12, 12, 12, 10, 12, 18, 16, 16, 16, 20, 12, 12, 12, 18, 18, 18, 11, 13, 16, 14, 12, 18, 16, 16, 16, 12, 16, 13, 16, 13, 12, 16, 16, 8, 18, 13, 18, 12, 16, 13, 16, 16, 16, 14, 18, 18, 16, 12, 13, 12, 13, 16, 13, 12, 12, 16, 21, 12, 16, 13, 13, 16, 13, 18, 13, 14, 16, 13, 9, 18, 16, 12, 12, 20, 12, 12, 16, 12, 18, 13, 16, 13, 12, 13, 13, 5, 13, 14, 11, 16, 12, 20, 5, 18, 14, 16, 13, 12, 18, 12, 18, 12, 14, 14, 16, 18, 14, 11, 10, 16, 12, 13, 13, 12, 21, 12, 13, 16, 16, 12, 12, 14, 14, 18, 12, 16, 12, 13, 16, 18, 13, 18, 12, 13, 21, 16, 13, 12, 9, 14, 16, 13, 16, 14, 12, 12, 12, 13, 16, 12, 16, 13, 12, 12, 12, 16, 16, 14, 12, 13, 18, 12, 16, 18, 16, 18, 13, 12,…
$ exper <int> 45, 25, 27, 42, 41, 26, 11, 15, 32, 12, 8, 40, 43, 23, 19, 33, 20, 30, 0, 5, 37, 32, 5, 5, 20, 7, 41, 32, 24, 20, 36, 7, 40, 29, 3, 13, 28, 10, 42, 37, 49, 20, 46, 43, 47, 47, 26, 21, 33, 6, 36, 39, 25, 22, 34, 16, 4, 13, 35, 12, 10, 20, 26, 17, 25, 26, 10, 13, 4, 14, 6, 11, 46, 7, 8, 25, 47, 5, 49, 20, 29, 10, 52, 22, 25, 5, 30, 0, 27, 23, 39, 8, 19, 11, 22, 25, 40, 6, 35, 39, 28, 45, 41, 30, 24, 30, 2, 6, 35, 49, 10, 15, 26, 20, 41, 14, 19, 22, 34, 4, 22, 15, 7, 5, 21, 36, 19, 29, 0, 16, 52, 4, 27, 31, 1, 21, 43, 35, 37, 5, 11, 8, 19, 29, 38, 17, 13, 36, 35, 31, 22, 29, 29, 53, 36, 5, 30, 40, 35, 34, 44, 37, 30, 33, 46, 30, 45, 43, 36, 21, 12, 47, 34, 3, 5, 32, 46, 1, 35, 15, 41, 12, 31, 20, 40, 38, 13, 13, 29, 14, 24, 46, 12, 33, 31, 8, 11, 4, 26, 39, 17, 27, 14, 33, 7, 27, 4, 24, 19, 3, 24, 13, 25, 7, 23, 3, 20, 9, 21, 12, 7, 28, 28, 42, 42, 12, 14, 38, 39, 27, 28, 10, 17, 32, 7, 11, 19, 25, 14, 33, 33, 24, 50, 6, 12, 16, 5, 25, 22, 14, 20, 32, 13, 45, 36, 38, 33…
$ faminc <int> 0, 45351, 91946, 48370, 10000, 151308, 110461, 0, 67084, 14000, 0, 25000, 0, 0, 50808, 63169, 162, 134370, 0, 77280, 80337, 23472, 18200, 86400, 0, 0, 0, 30669, 0, 0, 49400, 20400, 50000, 2395, 0, 0, 0, 17544, 0, 50169, 9000, 10000, 45993, 83258, 24019, 51475, 19906, 9904, 95000, 40002, 0, 0, 15100, 530, 5000, 25000, 30000, 3090, 0, 33162, 0, 58540, 5000, 0, 20000, 0, 34000, 0, 115500, 66000, 0, 0, 50500, 7700, 30000, 96056, 10799, 20000, 9599, 0, 32000, 0, 30000, 30003, 117240, 0, 112075, 117580, 66600, 30032, 25000, 0, 0, 90056, 24700, 104000, 0, 0, 1815, 33200, 0, 23886, 42992, 30000, 14240, 0, 0, 28962, 24002, 25002, 60000, 100081, 13600, 0, 65112, 0, 0, 68015, 70000, 0, 96010, 22679, 42500, 50500, 38062, 135088, 20000, 1220, 21256, 0, 44549, 0, 31400, 52002, 0, 60000, 41748, 112429, 118570, 20002, 35000, 30002, 24600, 71510, 113000, 40405, 3300, 23000, 42940, 130422, 8000, 71300, 61685, 5, 0, 41050, 113113, 0, 0, 100001, 0, 25000, 106309, 91820, 0, 13769, 22920,…
$ female <int> 1, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 1, 0, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 1, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 1, 0, 1, 1, 1, 0, 0, 0, 1, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 1, 0, 1, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 1, 0, 0, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 1, …
$ metro <int> 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 0, 1, 1, 1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 1, 1, 0, 1, 1, 0, 1, 1, 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0, 1, 1, 0, …
$ midwest <int> 0, 1, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 1, 0, 1, …
$ south <int> 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, …
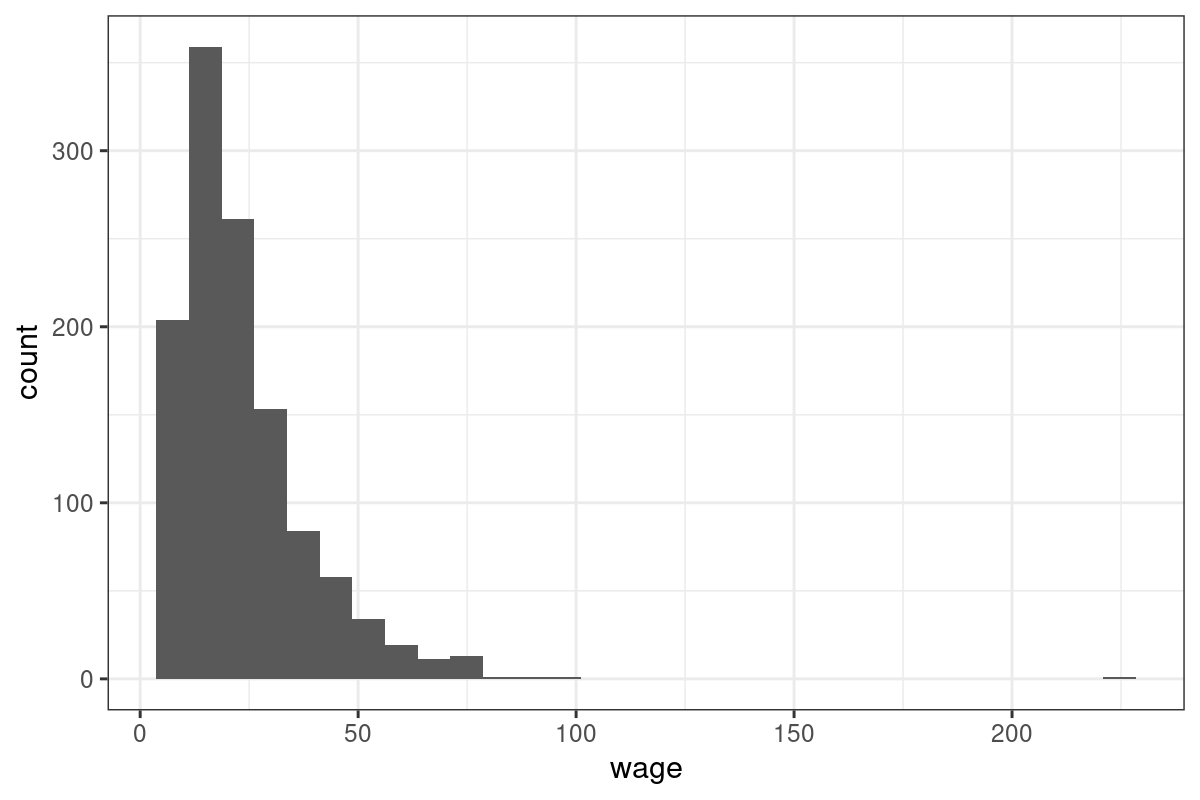
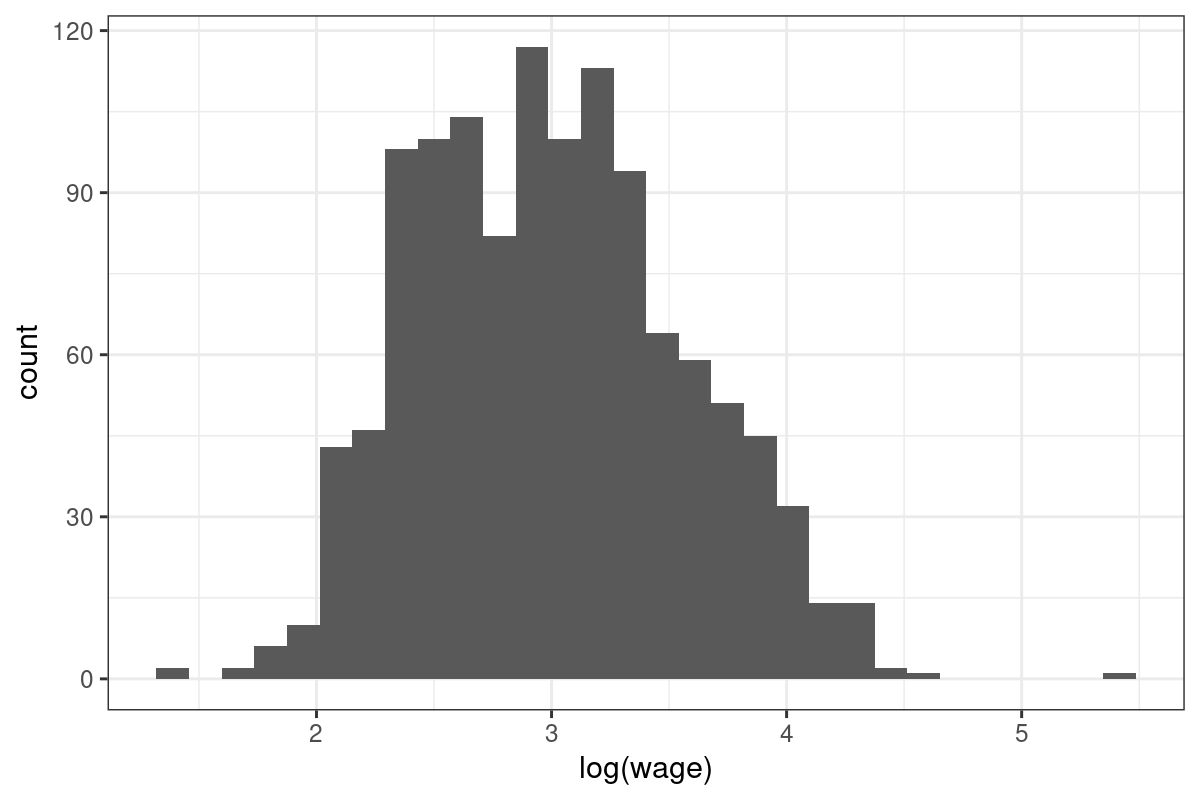
$ wage <dbl> 44.44, 16.00, 15.38, 13.54, 25.00, 24.05, 40.95, 26.45, 30.76, 34.57, 20.67, 21.50, 11.77, 24.35, 55.00, 11.00, 57.70, 58.00, 5.75, 11.50, 33.65, 10.50, 14.50, 8.25, 8.65, 13.00, 17.00, 16.33, 7.20, 10.00, 60.83, 17.80, 30.38, 12.50, 49.50, 23.08, 28.95, 9.20, 29.00, 15.00, 14.70, 23.66, 8.00, 13.66, 15.00, 10.50, 7.75, 11.64, 37.03, 9.15, 17.00, 15.42, 43.28, 19.23, 16.00, 31.92, 30.00, 9.50, 17.30, 17.92, 12.50, 16.00, 13.71, 18.12, 26.00, 19.30, 10.00, 38.08, 12.00, 42.16, 28.40, 9.75, 36.06, 18.75, 10.50, 33.65, 17.00, 24.47, 9.80, 22.72, 11.88, 11.00, 8.76, 24.02, 14.73, 13.33, 41.83, 15.83, 42.73, 31.25, 17.00, 15.00, 25.00, 34.63, 9.50, 16.60, 23.00, 16.48, 48.08, 17.23, 34.60, 27.68, 21.00, 16.83, 36.06, 57.70, 11.50, 8.00, 14.00, 21.63, 19.23, 19.44, 10.85, 23.55, 30.00, 40.47, 11.77, 27.50, 22.00, 9.48, 15.38, 17.30, 17.40, 21.63, 27.47, 18.00, 28.83, 20.00, 9.00, 21.00, 64.09, 14.00, 18.00, 11.35, 6.17, 26.22, 18.00, 18.65, 45.01, 11.00, 44.51, 32.00, 43.2…
$ west <int> 1, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, …